32 Expanding Data
Values can often be missing in your data, and sometimes entire observations are absent too. In this chapter, you’ll learn how to complete your dataset with these missing observations. You’ll add observations with zero values to counted data, expand time series to a full sequence of intervals, and more!
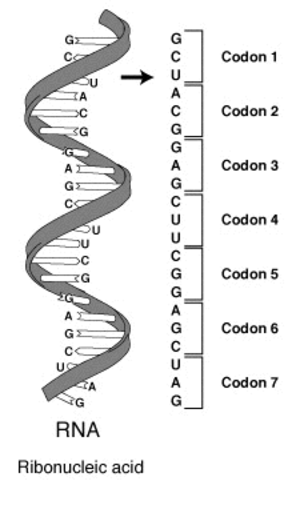
32.1 Letters of the genetic code
The basic building blocks of RNA are four molecules described by a single letter each: adenine (A), cytosine (C), guanine (G), and uracil (U). The information carried by an RNA strand can be represented as a long sequence of these four letters. To read this code, one has to divide this chain into sequences of three letters each (e.g.** GCU, ACG**, …). These three letter sequences are known as codons. The concept is illustrated in the image below.
Instructions 100 XP
- Create a tibble with three columns called
letter1,letter2, andletter3that holds all possible combinations of the vector letters usingexpand_grid(). - Use the
unite()function from chapter one to merge these three columns into a single column named codon. Use an empty string as the separator.
ex_019.R
letters <- c("A", "C", "G", "U")
# Create a tibble with all possible 3 way combinations
codon_df <- expand_grid(
leter1 = letters,
leter2 = letters,
leter3 = letters
)
codon_df
letters <- c("A", "C", "G", "U")
# Create a tibble with all possible 3 way combinations
codon_df <- expand_grid(
letter1 = letters,
letter2 = letters,
letter3 = letters
)
#
codon_df %>%
# Unite these three columns into a "codon" column
unite("codon", c(letter1, letter2, letter3),
sep = ''
)32.2 When did humans replace dogs in space
You already know that in the early days of spaceflight, the USSR was testing rockets with dogs. You now wonder when exactly humans started replacing dogs on space flight missions. You’ve been given a dataset space_df with the number of both dogs (compiled by Duncan Geere) and humans in space per year from 1951 till 1970 (collected from Wikipedia).
Your goal is to create a plot that shows you the number of individuals sent into space per species. Before you can create this plot, you’ll first have to introduce zero values for missing combinations of year and species.
Load dplyr and ggplot2 packages.
Instructions 100 XP
- Create
full_df, a tibble with all unique combinations of the variables year (from 1951 to 1970) andspecies("Human"and"Dog"). - Perform a
right_join()betweenspace_dfandfull_dfon theyearand species columns. - Use the ggplot() function to create a line plot of n_in_space over year, colored by species.
- Use the replace_na() function to overwrite NA values in the n_in_space column with zeros.
ex_020.R
# Create a tibble with all combinations of years and species
full_df <- expand_grid(
year = 1951:1970,
species = c("Human", "Dog")
)
space_df %>%
# Join with full_df so that missing values are introduced
right_join(full_df, by = c("year", "species")) %>%
# Create a line plot with n_in_space over year, color by species
ggplot(
aes(
x = n_in_space,
y = year,
group = species,
color = species
)
) +
geom_line()
# Create a tibble with all combinations of years and species
full_df <- expand_grid(
year = 1951:1970,
species = c("Human", "Dog")
)
space_df %>%
# Join with full_df so that missing values are introduced
right_join(full_df, by = c("year", "species")) %>%
# Overwrite NA values for n_in_space with 0L
replace_na(list(n_in_space = 0L)) %>%
# Create a line plot with n_in_space over year, color by species
ggplot(aes(x = year, y = n_in_space, color = species)) +
geom_line()32.3 Finding missing observations
You’re an inspector at a nuclear plant and have to validate whether every reactor has received its daily safety check over the course of a full year. The safety check logs are in reactor_df, a data frame with columns date, reactor, and check.
Two vectors, dates and reactors, with all dates of the year and reactors at the plant respectively have been created for you. You’ll use the combination of the expand_grid() and anti_join() functions to find dates where particular reactors were not checked.
Load dplyr package.
Instructions 100 XP
- Use the
expand_grid()function to create a tibble holding all combinations of the variables date and reactor. Use the dates and reactors vectors created for you. - Perform an anti-join between f
ull_dfandreactor_dfon the date and reactor columns.
ex_021.R
# Create a tibble with all combinations of dates and reactors
full_df <- expand_grid(
date = dates,
reactor = reactors
)
# Find the reactor - date combinations not present in reactor_df
full_df %>%
anti_join(reactor_df, by=c("date", "reactor"))32.4 Completing the Solar System
You have been given a data frame (planet_df) from the devstronomy project with the number of moons per planet in our Solar System. However, Mercury and Venus, the two moonless planets, are absent. You want to expand this dataset using the complete() function and a vector planets that contains all eight planet’s names.
Instructions 100 XP
- Complete the
planetvariable using theplanetsvector. - Replace
NAvalues in then_moonsvariable with0Lvalues.
ex_022.R
planets = c(
"Mercury",
"Venus",
"Earth",
"Mars",
"Jupiter",
"Saturn",
"Uranus",
"Neptune"
)
planet_df %>%
complete(
# Complete the planet variable
planet = planets,
# Overwrite NA values for n_moons with 0L
fill= list(n_moons = 0L)
)32.5 Zero Olymoic medals
Since 1896, athletes from all over the world have been competing in the modern Olympic games. You’ve been given a dataset (medal_df) with observations for all medals won by athletes from the 10 most successful countries in Olympic history. You want to create a visual with the number of medals won per country (team) per year. However, since not all countries won medals each year, you’ll have to introduce zero values before you can make an accurate visual.
 Load
Load ggplot2 and dplyr. In step 2 and 3 the scale_color_brewer() function is used to color lines in the plot with a palette that makes it easier to distinguish the different countries.
Instructions 100 XP
- Count the number of medals won per team and year.
- Use
ggplot()to create a line plot withn_medalsoveryear, colored by team. - Complete the
teamandyearvariables, replaceNAvalues in then_medalscolumn with zeros.
ex_023.R
medal_df %>%
# Count the medals won per team and year
count(team, year, name = "n_medals")
medal_df %>%
# Count the medals won per team and year
count(team, year, name = "n_medals") %>%
# Plot n_medals over year, colored by team
ggplot(
aes(
x = year,
y = n_medals,
group = team,
color = team
)
) +
geom_line() +
scale_color_brewer(palette = "Paired")
medal_df %>%
# Count the medals won per team and year
count(team, year, name = "n_medals") %>%
# Complete the team and year variables, fill n_medals with zeros
complete(
team,
year,
fill = list(n_medals = 0)
) %>%
# Plot n_medals over year, colored by team
ggplot(aes(x = year, y = n_medals, color = team)) +
geom_line() +
scale_color_brewer(palette = "Paired")32.6 Creating a sequence with full_seq()
The full_seq() function will look for the minimal and maximal values inside the vector you pass it and will then generate a full sequence of numbers with a fixed period in between them. When used inside the complete() function, full_seq() is a handy tool to make sure there are no missing observations in your data. Before combining these two functions you’ll generate a few sequences with full_seq() on its own to get the hang of this function.
Instructions 100 XP
- Use
full_seq()to create a sequence with all years from 2020 till 2030. - Use full_seq() to create a sequence with all decades from 1980 till 2030.
- Use full_seq() to create a sequence with all dates in 1980 using the outer_dates vector.
ex_024.R
# Generate all years from 2020 to 2030
years <- full_seq(c(2020, 2030), period = 1)
years
# Generate all decades from 1980 to 2030
decades <- full_seq(c(1980, 2030), period = 10)
decades
outer_dates <- c(as.Date("1980-01-01"), as.Date("1980-12-31"))
# Generate the dates for all days in 1980
full_seq(outer_dates, period = 1)32.7 The Cold War’s hottest year
In October 1962, during the Cuban missile crisis, the world came close to a full scale nuclear war. Throughout 1962, the USA, USSR, and France together detonated a record 178 nuclear bombs for military power display and research. You’ve been given a sample of the Nuclear Explosion Database (NEDB) for that year (cumul_nukes_1962_df) with an observation for each date on which a bomb was detonated. The total_bombs variable contains the cumulative number of bombs detonated by a country up to that point in time.
You’ll complete the dataset to hold the full sequence of dates, and visualize the total number of bombs per country over time. You’ll also use the fill() function from Chapter One to impute missing values.
Load dplyr and ggplot2 packages.
Instructions 100 XP
- Complete the dataset so that for each
countrythere is an observation of each date using thefull_seq()function.
ex_026.R
cumul_nukes_1962_df %>%
# Complete the dataset
complete(
country,
date = full_seq(date, period = 1)
)
cumul_nukes_1962_df %>%
# Complete the dataset
complete(country, date = full_seq(date, period = 1)) %>%
# Group the data by country
group_by(country) %>%
# Impute missing values with the last known observation
fill(total_bombs)
cumul_nukes_1962_df %>%
# Complete the dataset
complete(country, date = full_seq(date, period = 1)) %>%
# Group the data by country
group_by(country) %>%
# Impute missing values with the last known observation
fill(total_bombs) %>%
# Plot the number of bombs over time, color by country
ggplot(
aes(
x = date,
y = total_bombs,
group = country,
color = country
)
) +
# These two lines will mark the Cuban Missile Crisis
geom_rect(
xmin = as.Date("1962-10-16"),
xmax = as.Date("1962-10-29"),
ymin = -Inf,
ymax = Inf,
color = NA) +
geom_text(
x = as.Date("1962-10-22"),
y = 15, label = "Cuban Missile Crisis",
angle = 90,
color = "white"
) +
geom_line()32.8 Olympic medals per continent
You want to compare Olympic performance of athletes per continent over time, both on the winter and summer Olympics. You’ve been given a dataset medal_df with the average number of medals won per participant of each continent since 1928 . You’ll complete this data to introduce zero values for years where a continent did not win any medals.
The ggplot2 package has been pre-loaded for you.
Instructions 100 XP
Complete the dataset so that each continent has a
medals_per_participantvalue at each Olympic event. Missing values should be filled with zeros.Nest the season and year variables using the
nesting()function, since the summer and winter Olympics don’t occur in the same years.Use
ggplot()to create a line plot with themedals_per_participantper year, color the plot by continent.
ex_026.R
medal_df %>%
# Give each continent an observation at each Olympic event
complete(
continent,
nesting(season, year),
fill = list(medals_per_participant = 0L)
) %>%
# Plot the medals_per_participant over time, colored by continent
ggplot(
aes(
x = year,
y = medals_per_participant,
group(continent),
color = continent
)
) +
geom_line() +
facet_grid(season ~ .)32.9 Tracking a virus outbreak
You’re a doctor in a remote village confronted with a virus outbreak. You have been collecting data on when your patients got infected and recovered in a data frame named patient_df. Your goal is to create a visual with the number of sick patients over time. You’ll first have to reshape the data so that you can count the number of sick patients per day.
The data frame has three columns: patient, infected, and recovered. The dplyr and ggplot2 packages have been pre-loaded for you.
Instructions 100 XP
Pivot the
infectedand recovered columns to long format, the old column names should go in thestatusvariable, the values todate.Group the data by patient and then complete the date column so that each date between infection and recovery is added using the
full_seq()column. At the end, ungroup the data.
ex_027.R
patient_df %>%
# Pivot the infected and recovered columns to long format
pivot_longer(
-patient,
names_to = "status",
values_to = "date"
)
patient_df %>%
# Pivot the infected and recovered columns to long format
pivot_longer(-patient, names_to = "status", values_to = "date") %>%
select(-status) %>%
# Group by patient
group_by(patient) %>%
# Complete the date range per patient using full_seq()
complete(date = full_seq(date, period = 1)) %>%
# Ungroup the data
ungroup()
patient_df %>%
# Pivot the infected and recovered columns to long format
pivot_longer(-patient, names_to = "status", values_to = "date") %>%
select(-status) %>%
# Group by patient
group_by(patient) %>%
# Complete the date range per patient using full_seq()
complete(date = full_seq(date, period = 1)) %>%
# Ungroup the data
ungroup()32.10 Counting office occupants
Imagine you’re an office facility manager and want to know how many people are present throughout the day. You’ve installed a sensor at the entrance that counts the number of people entering and leaving the building. The sensor sends an update at the end of every 20 minute time slot if at least one person passed.
To create a dataset ready for visualization, you’ll combine the different techniques you’ve learned so far.
Load dplyr and ggplot2 packages.
Instructions 100 XP
- Complete the time variable by using the
seq()function to create a sequence between the min and max values with an interval of “20 min”. Fill NA values of enter and exit with 0L.
ex_028.R
sensor_df %>%
# Complete the time column with a 20 minute interval
complete(time = seq(min(time), max(time), by = "20 min"),
fill = list(enter = 0L, exit = 0L)) %>%
# Calculate the total number of people inside
mutate(total_inside = cumsum(enter + exit)) %>%
# Pivot the enter and exit columns to long format
pivot_longer(enter:exit, names_to = "direction", values_to = "n_people") %>%
# Plot the number of people over time, fill by direction
ggplot(
aes(
x =time,
y = n_people,
fill = direction
)
) +
geom_area() +
geom_line(aes(y = total_inside))